小社が立地する熊本にある熊本大学とパートナーシップ協定を結んでいるタイの大学研究者によるライフサイエンス分野の連載(4 回)をお届けします。
The Stealthy Factor: sfRNA Orchestrates Infection

Sarin Chimnaronk
Laboratory of RNA Biology
Institute of Molecular Biosciences
Mahidol University
Abstract
Flaviviruses contain a single-stranded positive-sense RNA genome that serves as the template for both viral replication and translation. During the replication of the viral genomic RNA (gRNA), uncapped and decapped gRNAs become the target for cellular RNA degradation by the 5’-to-3’ exoribonuclease XRN1. However, the 3’ untranslated region (UTR) of gRNA is highly resistant to XRN1 and remains accumulated as a noncoding subgenomic flavivirus RNA (sfRNA) in infected cells. sfRNA is implicated in virus-induced cytopathicity and pathogenicity in vertebrate hosts. In mosquitoes, sfRNA plays a crucial role in overcoming the mosquito midgut barrier and virus accumulation in the saliva. While the functions of sfRNA have been extensively studied, the accurate molecular mechanisms by which sfRNA facilitates viral pathogenesis remain incompletely understood. This review summarizes the current knowledge on the mechanisms of sfRNA, with an emphasis on its intracellular interactions, and discusses the prospects for its applications in drug and vaccine development.
Introduction of sfRNA
Flaviviruses include many of the most prevalent viral scourges known to humanity, such as dengue virus (DENV), Zika virus (ZIKV), West Nile virus (WNV), Kunjin virus (KUNV), yellow fever virus (YFV), Japanese encephalitis virus (JEV), tick-borne encephalitis virus (TBEV), St. Louis encephalitis virus (SLEV), and Murray Valley encephalitis virus (MVEV): a total of 53 species are listed by the International Committee on Taxonomy of Viruses as of 2017 1). These viruses are transmitted by arthropod vectors, typically mosquitoes and ticks. DENV alone causes more than 100 million infections annually worldwide 2). The infection with flavivirus can cause a spectrum of diseases, ranging from mild febrile illnesses to severe hemorrhagic fevers and neurological complications. Despite the high disease burden, there are still no approved drugs for most flaviviral diseases. Flaviviruses are spherical enveloped particles of approximately 50 nm in diameter that contain a single-stranded, positive-sense RNA molecule of 10.12 kb long. The gRNA has a type I cap at the 5’ end but lacks a poly(A) tail at the 3’ terminus. The gRNA consists of a single open reading frame (ORF), which encodes three structural proteins (C, prM, and E) and seven non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (see Figure) 3). The viral polyprotein is cleaved by viral and host proteases into the mature proteins. The flavivirus genome is flanked by 5’ and 3’-UTRs, which are enriched with characteristic RNA secondary structures with crucial roles in replication and translation 4), 5). The 5’-UTR of flaviviruses spans around 100 nucleotides in length, whereas the 3’-UTR ranges from 400 to 700 nucleotides in length, depending on the virus species. A fascinating aspect of flavivirus biology, uncovered over the past two decades, is their production of a unique, noncoding RNA molecule known as subgenomic flavivirus RNA or sfRNA 6). Production of sfRNA by flaviviruses was first reported for MVEV 7). All known arthropod-borne flaviviruses produce 300–500-base-long sfRNA from the viral 3’-UTR by stalling the cellular 5’-to-3’ exoribonuclease XRN1 at a set of conserved pseudoknot-like RNA structures at the 5’ region of the 3’-UTR, resulting in the incomplete degradation of gRNA (see Figure) 8)-10). It should also be noted that multiple sfRNA isoforms with different sizes can be concurrently generated due to stalling of XRN-1 at different structural RNA elements in the 3’-UTR 11).
To date, studies on the functions of sfRNA have been actively conducted. WNV with impaired sfRNA production showed a significant reduction in virus-induced cytopathic effect in Vero cells and pathogenicity in mice 6). sfRNA alone did not induce apoptosis in BHK-21 cells, indicating that sfRNA must act under the condition of viral infection to promote virus-induced cytopathicity and apoptosis in mammalian cells 12). It has been demonstrated that both structural and non-structural proteins of flaviviruses exhibited pro-apoptotic activity, inducing cell death in various cell models 17). However, mutants of WNV and DENV deficient in the generation of sfRNA showed a drastic reduction in their ability to form plaques on Vero and BHK-21 cells, respectively. sfRNA-deficient viruses exhibited only 10% apoptosis, compared to 60-70% caused by the wild-type viruses 6), 12). It was also reported that sfRNA-deficient DENV failed to induce cleavage of caspase-3 and downstream cleavage of poly ADP-ribose polymerase (PARP), suggesting sfRNA triggers activation of caspase-3-dependent apoptotic pathways 12). Intriguingly, in contrast to vertebrate hosts, sfRNA suppressed apoptosis in ZIKV-infected mosquito cells, as required for virus accumulation in the saliva for productive viral transmission 13, 14). It is also noted that sfRNA in mosquito saliva is present in the extracellular vesicle (EV)-like particles 15). Unfortunately, the key pathway and mechanism underlying the discrepancy between induction and suppression of apoptosis in mammalian and mosquito cells remain poorly understood.
Besides cell death, the most investigated cellular pathway is the immunosuppression in hosts, as it is generally assumed that viruses should evade the innate immune response to facilitate efficient viral replication. Indeed, the DENV serotypes 2 (DENV-2) PR-2B strains that emerged during the epidemic in Puerto Rico in 1994 contained variants in the 3’-UTR, which significantly increased the level of sfRNA production per copy of gRNA than that of the DENV-2 PR-1 strains before the endemic 16). Epidemic strains also induced lower expression of interferon-beta (IFN-β) during infection in human hepatocellular carcinoma HuH-7 cells and primary monocytes. Moreover, transfection of in vitro transcribed sfRNA from PR-2B strain into HuH-7 cells together with poly(I:C), an immunostimulant, showed reduced expression of IFN-β compared to transfection of poly(I:C) alone and poly(I:C) with sfRNA from pre-epidemic strain. Unlike the apoptotic effect, these data support the rationale that the sfRNA molecule can directly suppress IFN-β, regardless of infection, and both the sequence and amount of sfRNA are key factors in the high attenuation of IFN-β expression. Also, transfection of an in vitro transcribed 5’-monophosphorylated DENV-2 3’-UTR into HuH-7 cells before infection by DENV-2 could abrogate the expression of both IFN-β and IFN-λ1 genes, as well as IFN-stimulated genes (ISGs) of ISG15 and myxoma resistance 1 (MX1), as measured by qRT-PCR 15). At present, it is generally accepted that sfRNA inhibits the induction of the type I IFN of IFN-β and potentially IFN-λ to enhance infectivity 18).
Remarkably, sfRNA is required for the pathogenicity of WNV in the mouse model. Mice infected with sfRNA-deficient viruses showed no sign of WNV-induced encephalitis and survived the infection, whereas mice challenged with the wild-type virus were 100% lethal at day 9 6). The difference in viral load in the mouse brain was not obvious, indicating sfRNA is not essential for viral replication and spread in vivo. Thus, the lack of neuropathogenicity is conceivably associated with the loss of sfRNA, which prevents the virus from inhibiting type I interferon and/or eliciting apoptosis. However, no proof was provided for the increased apoptosis or the impairment of immune function in the infected mouse brain. It also requires further study to investigate if the sfRNA-dependent pathogenicity in vertebrates can be extrapolated to other flaviviruses.
Physical interactions of sfRNA
Before we go any further, let's take a close look at the conserved RNA secondary structures in the flaviviral 3’-UTRs, using the DENV-2 UTR as an example (see Figure). The flaviviral 3’-UTR can be divided into 3 regions. From the 5’ end is the variable region containing duplicated structured stem-loop RNA elements (SLs). The number of SL can be varied in each flavivirus strain. More importantly, these SL structures can form the pseudoknot RNA secondary structure and fold into a unique, compact three-dimensional structure of a three-helix junction, which halts XRN1 and produces sfRNA 20). These RNA domains are also called exoribonuclease-resistant RNAs (xrRNAs). The three 5’ nucleotides upstream of SL are protected from XRN1 degradation by base pairing within a ring-like structure of xrRNA. Disruption of these base pairs by mutations abolished the ability to resist XRN1 degradation 20). Since the configuration of the three-helix junction formed in xrRNAs has not been previously observed and cannot be classified into the three known types of three-way junctions, flaviviruses have intentionally acquired the xrRNA module and XRN1-resistant mechanism over a long period of evolution to produce sfRNA.
The middle domain of flaviviral 3’-UTR has a single or two similar dumbbell-shaped RNA structures (DB1 and DB2). Remarkably, the hairpin occupying the 3’ half of DB harbors the most significantly conserved sequence among flaviviruses, implying a fundamental function in the virus life cycle 21). At the 3’ region of the 3’-UTR, there is the longest terminal 3’-stem loop (3’ SL) preceded by a short hairpin (sHP). The apical loop of 3’ SL contains a highly conserved pentanucleotide of CACAG, which is required for viral replication 22). Hence, the unique and conserved RNA sequences and structures in the flaviviral 3’-UTR and sfRNA can be easily predicted to serve as a platform for the interactions with protein and protein complexes, so-called “protein sponge”, to control or fine-tune processes in the viral life cycle.
To comprehensively understand the action of sfRNA, several efforts have been made to identify the cellular proteins that interact with sfRNA in infection. Due to current advances in computer science, a large-scale in silico exploration of the global interaction network, known as the “interactome”, of sfRNA across multiple flaviviruses, including DENV, ZIKV, JEV, YFV, and WNV, was conducted using the catRAPID omics server 19). The results revealed five human RNA-binding proteins (RBPs), which were DDX1, NKRF, CSTF3, TRM1L, and NUFP2, that were predicted to interact with sfRNAs of all flaviviruses tested. The computational screening of protein-RNA interactions is a fascinating yet poorly experimentally validated area. It is hoped that the rapid progress of artificial intelligence (AI) will enable accurate and reliable prediction of interactions in the near future. Alternatively, the general method for identifying proteins interacting with the RNA of interest involves a combination of RNA affinity pull-down and mass spectrometry (AP/MS). An earlier study using AP/MS identified the Y-box-binding protein (YBX1) bound to two loops in the DENV-3 3’ SL 23). However, YBX-1 was not found in another AP/MS study with DENV-2 UTRs 24). This discrepancy in the published dataset for sfRNA-bound proteins is often observed in the AP/MS studies, likely due to the variation in the viral RNA sequences, affinity tag sequence, cell types, experimental conditions, and analysis procedure. It should also be noted that affinity pull-downs might not be performed in an environment mimicking viral infection. In particular, the use of uninfected cell lysate for the pull-down is likely to introduce significant bias by ignoring gene expression patterns, protein localization, and time course in infection. To date, there are very few interaction pairs of sfRNA and host proteins that have been characterized for their accurate recognition motifs and downstream molecular effects of interaction.
So far, the most characterized sfRNA interactions are enriched in the 5’ variable region of DENV sfRNA. As described above, XRN1 stalls by the ring-like configuration of xrRNAs and produces sfRNA. A previous study demonstrated that sfRNA sequesters XRN1 during DENV and KUNV infection to dysregulate host mRNA turnover, as observed for the accumulation of uncapped mRNAs and increased stability of host transcripts in HEK293T cells. However, it requires further testing in more cell types to generalize this finding. It is also unclear how the alterations in host mRNA half-lives would impact the regulation of interferon expression and cell death. A fascinating finding is the interaction of the regulators of cytoplasmic stress granules (SGs) with the variable region of DENV sfRNA. G3BP1, G3BP2, CAPRIN1, and USP10 are proteins localized to SGs, which were found to interact with the first xrRNA (SL-I in DENV-2) 24),25). SGs are cytoplasmic RNA granules that act as sites for the storage and/or degradation of mRNAs under cellular stress conditions that inhibit translation initiation. It was proposed that the binding of these SG proteins to DENV-2 sfRNA inhibited translation of two selected interferon-stimulated genes (ISGs) of IFITM2 and PKR to down-regulate the antiviral activity of IFN-β. However, this interaction of sfRNA with SG components was not reproducible in the DENV-3 strain and other flaviviruses 25) and was not found in another AP/MS study on ZIKV sfRNA 26). Lastly, the variable region of sfRNA from clinical isolates of DENV-2 was shown to interact with the ubiquitin ligase tripartite motif-containing protein 25 (TRIM25) and prevent deubiquitination of TRIM25 to counteract RIG-Iinduced IFN-β expression. Again, the TRIM25 interaction was proven only in specific DENV-2 strains and not identified in other AP/MS studies with DENV-2 and ZIKV 24), 26). Interestingly, TRIM25 possesses robust RNA-binding activity in its C-terminal PRY/SPRY domain and also forms the complex with SG proteins, including G3BP1 and USP10 27). Because evidence for interactions with the variable region of sfRNA was mainly provided by the pull-down experiments with cell lysate, it should therefore not be simple to argue the direct binding of those proteins to the sfRNA sequences and structures.
Our study and other studies identified DDX6, a DEAD-box RNA helicase implicated in the formation or function of processing bodies (PBs) and SGs, as an interactor of the dumbbell RNAs in sfRNA 21), 24), 26). We first pinpointed the DDX6-binding site to the completely conserved hairpin at the 3’ half of DB2 and measured their affinity (Kd) to be ~8 nM 21). This interaction has been confirmed in DENV-1–4, ZIKV, and WNV in vertebrate cells and even in mosquito cells 14). Our results showed that sequestering DDX6 by DB2 resulted in host cell cycle arrest in the G1 phase 21). Intriguingly, DDX6 was found to be a new regulator of the antiviral IFN response 28). Further studies are clearly required to identify the molecular events downstream of DDX6 sequestration by sfRNA, which may advance our understanding of the innate immunomodulatory activity of sfRNA.
The last domain of sfRNA contains a 3’ SL element. Our previous study revealed that the conserved pentanucleotide in the apical loop of 3’ SL is recognized and bound to the viral NS5 RNA-dependent RNA polymerase (RdRp) (see Figure) 22). This interaction is indispensable for the replication of gRNA. The binding of viral NS5 to sfRNA was also described in a recent AP/ MS study using ZIKV-infected cell lysate 29). Notably, it was suggested that the NS5-bound sfRNA complex interacts with signal transducer and activator of transcription 1 (STAT1), leading to a reduction in phosphorylation and nuclear translocation of STAT1 to block type I and III IFN signaling 29). This interaction currently offers the most comprehensive insight into the mechanism by which sfRNA antagonizes the interferon (IFN) response.
Implication of the flavivirus live-attenuated vaccine
The strong association between sfRNA production and the pathogenicity of flaviviral diseases emphasizes its clinical importance and identifies it as a compelling candidate for novel antiviral intervention strategies such as a live-attenuated vaccine. A tetravalent DENV vaccine, named TV003, that is currently in phase 3 clinical trials, was developed by introducing a 30-nucleotide deletion (Δ30) in the 5’ hairpin of DB2 in the 3’-UTR (review in 30)). The phase I clinical trial demonstrated that a single dose of TV003 induced a robust, balanced neutralizing antibody response against all four serotypes of DENV, with no subjects experiencing any dengue-like illness 31). Intriguingly, the DENV Δ30 vaccine strain decreased the accumulation of sfRNA in infected human cells 32). Later, it was reported that the deletion of only the last 10 nucleotides (Δ10) of Δ30 was sufficient and essential to make ZIKV attenuated in the mouse model 33). Moreover, the 3’ region of the Δ30 and Δ10 deletion always overlaps (5-6 nucleotides) with the DDX6-binding site in DB2, which was shown that mere deletion of this overlapped sequence abolished DDX6-binding capacity 21). Collectively, these disparate findings converge to support a model of flavivirus attenuation in which DDX6 is central to sfRNA stabilization and the regulation of IFN signaling, which awaits further investigations.
Conclusions and future directions
The structured noncoding sfRNA highlights the evolutionary innovation of flaviviruses, serving as a streamlined and potent mechanism for host cell manipulation. Accumulating evidence has firmly established sfRNA as a key regulator of host immune modulation and viral pathogenicity. Here, we propose that functional interactions of sfRNA with NS5 and DDX6 are a pivotal mechanism underlying these effects. DDX6 is an essential and evolutionarily conserved protein fundamental for mRNA translation control, storage, and degradation. Further studies are clearly required to clarify how sfRNA-mediated sequestration of DDX6 contributes to the broader reprogramming of RNA metabolic pathways. Also, further studies to provide insights into the three-dimensional architecture of sfRNA in complex with its interacting protein partners are important for the rational development of vaccines and antiviral agents designed to block sfRNA biogenesis or disrupt its functional interactions. Such structural and mechanistic insights will not only deepen our understanding of flavivirus-host interactions but also pave the way for targeted therapeutic strategies that exploit vulnerabilities in sfRNA-mediated immune evasion.
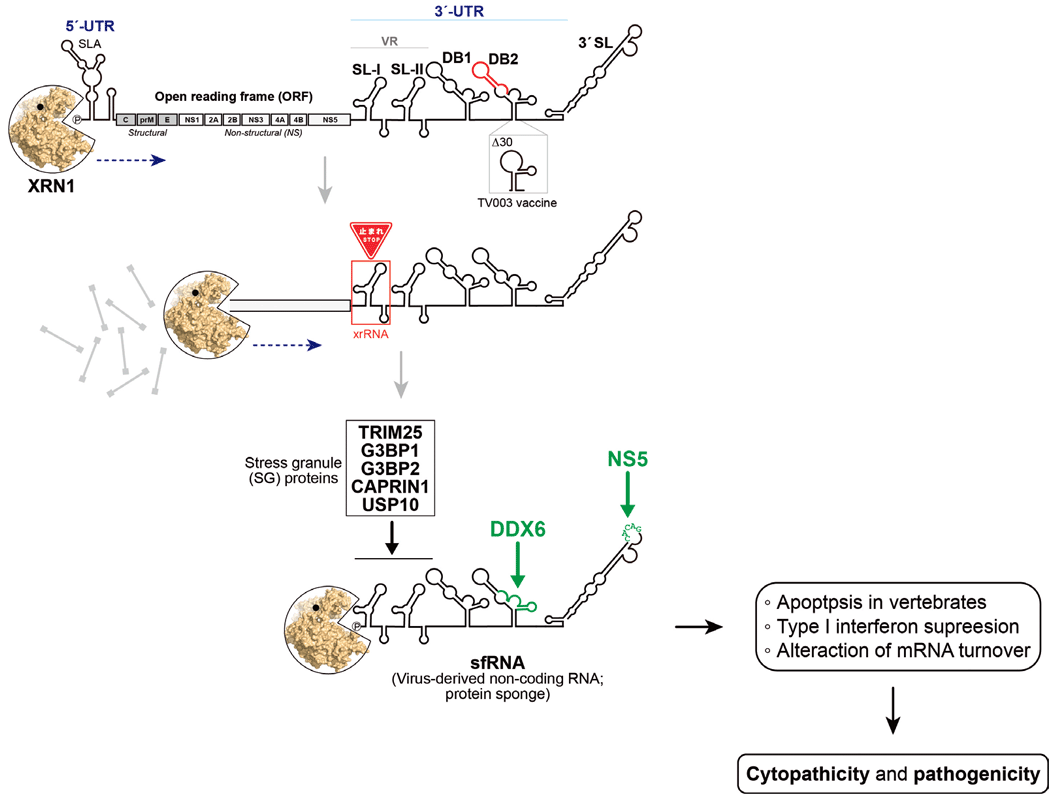
Schematic illustration of subgenomic flaviviral RNA (sfRNA) biogenesis and functions through XRN1 (Pacman)-mediated degradation of viral genomic RNA (gRNA). The representative genome of dengue virus serotype 2 (DENV-2) is depicted, including its overall organization and conserved RNA structural elements within the 3’-untranslated region (3’-UTR) ̶ notably stem loops (SL), dumbbells (DB), short hairpins, and the 3’-terminal stem loop (3’ SL). The SL-I element, also referred to as xrRNA, adopts a three-dimensional conformation that effectively stalls XRN1-mediated degradation. Two protein-binding sites, identified in our previous studies, in DB2 and the 3’ SL are marked in green. sfRNA has been implicated in flavivirus-induced cytopathic effects and contributes to viral pathogenicity.
[Contact]
Sarin Chimnaronk, Ph.D.
ORCID ID: 0000-0001-6113-3681
Laboratory of RNA Biology,
Institute of Molecular Biosciences, Mahidol University.
25/25 Phutthamonthon 4 Road, Salaya, Nakhon Pathom 73170, Thailand.
Current research area: Viral RNA-host interaction, RNA virus replication, mRNA vaccine