Methionine-Selective Protein Bioconjugation
 |
Yvonne D. Hall Department of Chemistry, North Carolina State University Undergraduate Researcher |
 |
Hisham M. El-Shaffey Department of Chemistry, North Carolina State University Graduate Student |
 |
Elizabeth J. Gross Department of Chemistry, North Carolina State University Undergraduate Researcher |
 |
Jun Ohata Department of Chemistry, North Carolina State University Assistant Professor |
Abstract
Despite significant advances of protein bioconjugation strategies targeting a variety of amino acid residues, chemical tagging of methionine residues have been extremely limited.
Recent approaches leverage unique reactivity of the thioether group of the methionine residue through redox and alkylation reaction pathways with the suppression of cross reactivity toward other nucleophilic amino acid side chains. This Review Article highlights the development and applications of the three distinct chemical probes recently reported for selective modification of methionine residues of natural proteins.
1. Introduction
Chemical functionalization of natural proteins has become an essential technology in a wide variety of protein-based research fields and applications. Protein modification technology provides opportunities for the increase of stability, addition of new functionality, and structural changes. One important aspect of protein modification is the development of therapeutic agents; for example, antibody-drug conjugates(ADCs)are target-specific proteins covalently labeled with potent small molecule-based drugs. 1) Traditionally, protein modifications target highly nucleophilic amino acid sites such as lysine(primary amine)and cysteine(thiol). However, because of lysine's high abundance, targeted reactions are prone to heterogenous labeling of multiple sites, impairing reproducibility and structural/functional characterization. In this context, cysteine modification benefits from its lower abundance, but the formation of the disulfide bonds and other various post-translational processes greatly decrease their accessibility. Thus, a significant need for development of novel protein modification technologies to target different amino acid residues remains.
Due to its unique reactivity and low natural abundance, methionine offers the opportunity for the selective labeling of proteins. The sulfur atom of the methionine provides its functional diversity in living systems such as sulfoxide formation under the oxidative stress and coordination to various metal ions. The scarcity and hydrophobicity of methionine is beneficial for the development of new site-specific labeling technologies, as those attributes limit the number of surface-exposed residues accessible for the modification process. Despite the promising attributes, however, selective chemical modification of methionine residues still remains underdeveloped, in part due to the presence of the plethora of other nucleophilic functional groups in proteins. Alkyl halide and cyanogen bromide are the classical examples of methionine-targeting reagents, though those approaches suffer from various issues such as harsh conditions, side reactions, and/or limited substrate scopes. 2)-4)
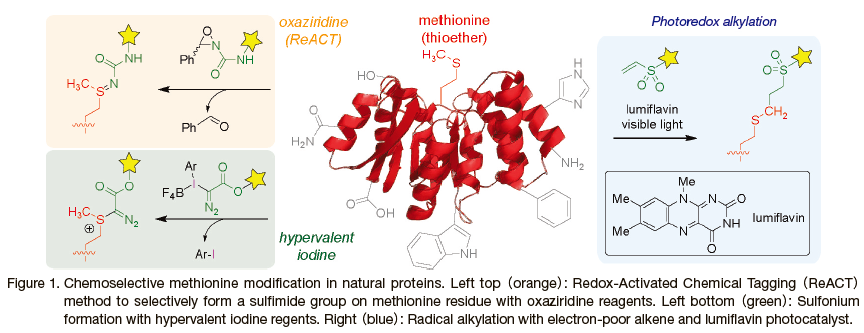
In this Review Article, we focus on three recently reported methionine-specific bioconjugation methods and their applications for biological studies(Figure 1). The seminal work of the modern methionine labeling strategies is the sulfimide formation reaction with the oxaziridine reagent termed Redox-Activated Chemical Tagging(ReACT) method developed by the Chang and Toste laboratories in 2017(Figure 1, left top in orange). 5) Inspired by the native biological oxidation of methionine thioether to methionine sulfoxide, this sulfimide-forming reaction involves the nucleophilic attack of the sulfur atom to the nitrogen atom of the oxaziridine labeling probe through release of a benzaldehyde group. In the following year, Gaunt and co-workers reported hypervalent iodine-based diazo sulfonium forming reaction on methionine residue, which introduces diazo ester group onto the sulfur atom through generation of the aryl iodide side product(Figure 1, left bottom in green). 6) More recently, Macmillan and co-workers demonstrated radical alkylation reaction of the methionine with electron-deficient alkene and a lumiflavin photocatalyst through the formation of the radical species on the terminal carbon of the methionine side chain(Figure 1, right in yellow). 7) The following sections will highlight the unique applications of each methionine labeling technology.
2. Oxaziridine reagents
2.1. First-generation oxaziridine for proteomics and imaging applications
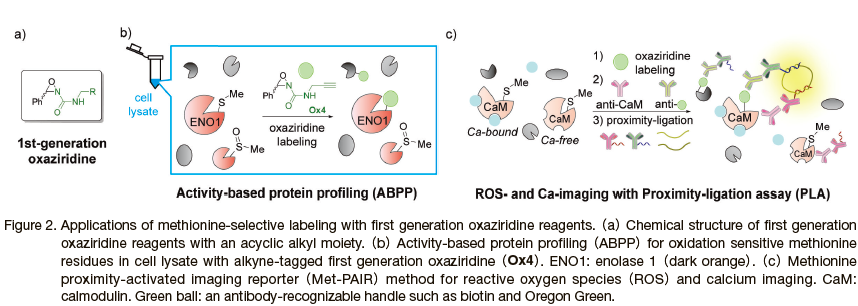
An early success of the Redox-Activated Chemical Tagging(ReACT)technique was in its application as a chemical probe for activity-based protein profiling(ABPP)to elucidate relationships between protein activity and the reversible oxidation of methionine residues, an emerging physiologically important class of redox post-translational modifications.5) One notable example of methionine oxidation is in the regulation of actin polymerization; Terman and co-workers demonstrated that actin assembly and disassembly are controlled by the oxidation and reduction of the Met44 and Met47 residues by MICAL, an intracellular flavoprotein monooxygenase. 8) The current understanding of the physiological roles played by methionine residue oxidation has lagged behind other amino acid oxidations such as cysteine, whose impacts on disease progression is well documented, 9) due to a lack of suitable chemical probes. In this context, ABPP provides a chemoproteomic approach that determines the functional state of proteins through chemical labeling. 10) As the oxaziridine labeling of a methionine residue cannot proceed on a methionine sulfoxide group, the labeling efficiency of a methionine residue on a protein in a biological sample would be inversely correlated to its oxidation status.
Indeed, owing to its high chemoselectivity for methionine and compatibility with cellular conditions, the ReACT method was successfully employed as an ABPP tool to mammalian cell(HeLa cells)lysates with varying doses of oxaziridine used to identify proteins with oxidatively sensitive methionine residues.
Validating oxaziridine-based ABPP and demonstrating applicability of the ReACT method to complex biomolecule mixtures, the proteomics results identified the actin Met44 and Met47 residues mentioned above at the lower dose levels of oxaziridine reagents. Importantly, the oxaziridine-based ABPP also identified the Met 169 residue of enolase, near the protein's active site, as a highly reactive target. Subsequent experiments with site-specific mutagenesis and kinetic measurement revealed that the oxidation of enolase Met 169 residue is a crucial modulator of enolase's enzymatic activity. This study demonstrates the practical utility of the oxaziridine reagents for understanding biological roles of methionine oxidation. 5)
The excellent stability and selectivity of oxaziridine reagents allow for their use in protein labeling in vivo, which has been utilized for calcium and reactive oxygen species(ROS)imaging together with proximity ligation assay(PLA)in a collaboration between the Chang, Miller, and Toste laboratories. 11) Reactivities of methionine residues in a particular protein can be significantly altered in response to cellular small molecules, and the reactivity alteration has been applied for calcium imaging through chemical labeling of calcium binding protein calmodulin(CaM, Figure 2c).
Because of the conformational change and exposure of methionine residues in the presence of calcium, the chemical labeling efficiency of the oxaziridine toward those methionine serves as a proxy for the amount of calcium in the biological samples such as cultured mammalian cells(e.g. HEK293T, Jurkat, and hippocampal neuron cells)as well as zebrafish. Although the methionine-selective reaction itself cannot function as a calcium reporter due to off-reactivity toward other methionine-containing proteins, PLA with antibodies against CaM and chemical tag(green ball in Figure 2c)as well as secondary antibodies bearing corresponding DNA strands provide fluorescence signals only from oxaziridine-labeled CaM through consecutive DNA hybridization processes(Figure 2c). Furthermore, taking advantage of the reactivity decrease of methionine residues upon oxidation(i.e. methionine sulfoxide formation)in actin, the dual labeling by methionine-selective oxaziridine compound and PLA technique can sense ROS levels in live cell samples. This technique termed methionine proximity-activated imaging reporter(Met-PAIR)is a unique application of chemoselective protein modification in living systems to visualize the intracellular amount of a biological analyte, which would not be easily accomplished by traditional protein labeling reagents such as iodoacetamide for cysteine residues due to their high cytotoxicity. More recently oxaziridines have been employed by researchers at Bristol-Myers Squibb for developing novel 18F PET imaging agents as well(Section 3). 12)
2.2. Second-generation oxaziridine for preparation of stapled peptides and antibody-drug conjugates
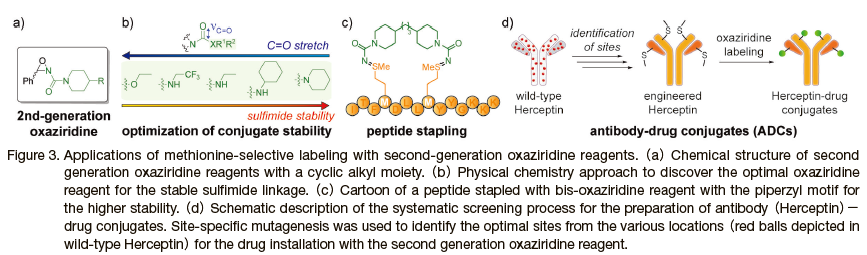
In order to modulate the stability of the modification linkage between proteins and installed functionalities, the tunability of the sulfimide linkage was investigated to facilitate wider adoption of the oxaziridine labeling technique. 13) Toste, Chang, and co-workers probed the physical parameters of a library of oxaziridine reagents, with varying functional groups attached to the urea moiety(Figure 3a, b). Among the various physical parameters of oxaziridine compounds with different electronic and steric functional groups including the urea's dipole moment(μ)and sulfimide's natural bond order(NBON), the vibrational constant of the urea C=O stretch(νC=O)yielded the best correlation with the experimental decomposition rate of sulfimide hydrolysis. In other words, the carbonyl stretching frequency provided a predictive platform for the design of next generation oxaziridine with the linkage stability. With piperidine as a substitution group, the next generation of oxaziridine displayed significantly increased sulfimide linkage stability. Stability comparisons, under physiologically relevant conditions(pH 7.4 and 37℃ in phosphate buffered saline)demonstrated that 59% of sulfimide linkages produced by the second-generation oxaziridine remained even after 5 days while sulfimide linkages produced by the first-generation oxaziridine had completely decomposed. As a further demonstration of the enhanced utility of the second-generation oxaziridine, bis-oxaziridine compounds were synthesized and reacted with several peptides to prepare a series of stapled peptides, and indeed the stapled peptides demonstrated improved cellular uptake with human embryonic kidney cell lines(HEK293T)using confocal microscopy and flow cytometry.
The second generation oxaziridine with the increased modification linkage stability was also utilized for production of antibody-drug conjugates(ADCs)in combination with site-specific mutagenesis(Figure 3d). 14) The study led by Wells and co-workers focused on the identification of the optimal modification site of anti-Her2 antibody(trastuzumab or Herceptin)through systemic approaches. Of the 95 potential mutation sites of antigen-binding fragment(Fab)of trastuzumab, 56 mutants were identified satisfying the ADC production criteria including high expression level through E. coli, high binding affinity, thermal stability, and efficient labeling with first generation oxaziridine reagents. Alongside the second generation oxaziridine reagents, further investigation revealed 4 sites on the Fab region can generate suitable conjugates with enhanced stability(> 85% conservation of the oxaziridine label after 3 days at 37 ℃).
Conjugation of the mutated antibody fragment with the synthetic peptide-based tubulin inhibitor monomethyl auristatin F(MMAF)produced potent ADCs toward breast cancer cell lines such as BT474-M1. By taking advantage of the structural homology between Fab and Fc regions of the antibody, the screening process was extended to discover the optimal conjugation sites in the Fc region as well. With the ideal modification sites identified for both Fab and Fc regions, mutants of the whole trastuzumab were produced(e.g. LC.T74M/ HC.S21M)and tested on multiple breast cancer cells lines and a BT474-xenograft mouse model, showing the promoted efficacy of the ADCs compared to the unmodified antibody and unconjugated MMAF. In addition to the practical success of this approach to generate the potent ADCs, this study also illustrated the potential utility of the methionine as bioconjugation sites by suppression of the nonspecific reactivity such as that of lysine and cysteine.
3. Applications of hypervalent iodine S-alkylation and photoredox alkylation
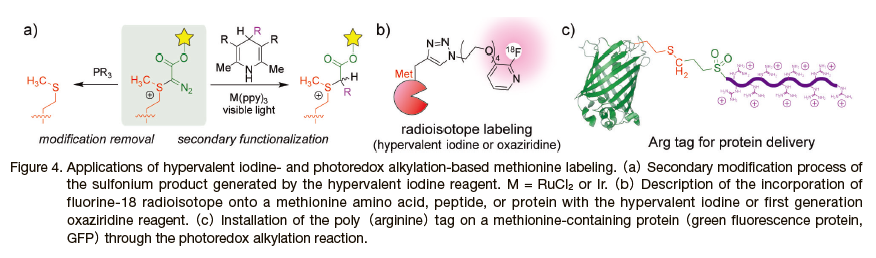
The versatile reactivities of the diazo sulfonium species formed by the hypervalent iodine reagent allowed for secondary functionalization on the methionine residue, including the subsequent radioisotope incorporation(Figure 4). 6) While the sulfonium product possesses remarkable stability in water(half-life > 100 h), the inherent reactivity of the diazo group enables attachment of various groups through a photocatalytic reduction as well as phosphine-based reduction to cleave the entire modification from the residue(Figure 4a). By leveraging the stability increase of the photocatalytically reduced sulfonium group, Bonacorsi and co-workers demonstrated successful incorporation of 18F radioisotope through copper-catalyzed azide-alkyne cycloaddition reaction. 12) Thus, the sulfonium linkage is of great use for the modulation of the stability and subsequent addition of a new functionality through the photoredox catalysis.
The photoredox alkylation reaction facilitates the incorporation of diverse chemical functionalities to a methionine residue of a protein(Figure 4c).7) Despite the generation of the putative methionine radical species through single-electron transfer(SET)process during the modification, this photoredox modification exhibited excellent functional group tolerability toward a variety of functional groups including primary amines,
azides, alkynes, and ketones. Conservation of native protein functions was confirmed on the alkyne-labeled enhanced green fluorescence protein(eGFP). The modified eGFP was amenable to the subsequent copper-catalyzed azide-alkyne reaction to append biotin and poly(arginine) tags, which displayed increased cellular uptake in the confocal microscope experiment(Figure 4c).
4. Conclusion
The three classes of methionine-selective chemical reagents developed for protein bioconjugation presented here possess potential for a broad range of applications largely due to rapid reaction kinetics. Although good selectivity has been achieved, particularly in the Redox-Activated Chemical Tagging(ReACT)method, combining approaches that can circumvent the problems of undesired side reactions, including cysteine-capping strategies with a maleimide reagent(hypervalent iodine)6 and use of inert atmosphere to curtail methionine oxidation(photoredox alkylation), can augment this selectivity even further. 7) Indeed, three independent research programs developed vastly different chemical approaches to form distinct product structures, reflecting the diverse chemical and biological reactivity of methionine: 1)sulfimide formation(oxaziridine labeling)/methionine sulfoxide(oxidation of methionine), 2)sulfonium formation(hypervalent iodine)/S-adenosyl methionine(SAM, S-alkylation of methionine), and 3)C-C bond forming modification(photoredox catalysis)/intact methionine. The diverse achievements of those chemical probes are indicative of their exceptional utility for the elucidation of undiscovered biological roles of methionine as well as the novel protein conjugates preparation.
| [ Contact ] |
Yvonne D. Hall North Carolina State University |
Jun Ohata (大畠潤) North Carolina State University |

