Select Your interested category.

Chromogenic Dyes
Trinder's Reagents
| Name | Code | CAS# | Chemical Structure | MW | Chromogen (With 4-AA,POD) |
|
|---|---|---|---|---|---|---|
| λmax | ε×104 | |||||
| TOOS | OC13 | 82692-93-1 | 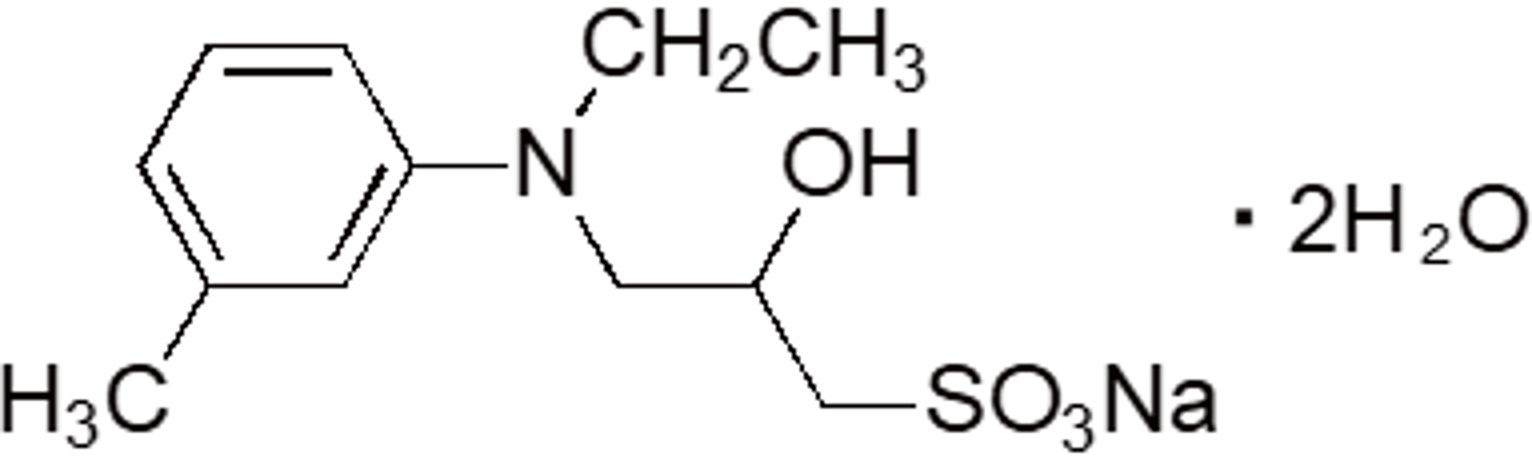 |
331.36 | 555 | 3.92 |
| TODB | OC22 | 1044537-70-3 |  |
423.46 | 550 | 3.80 |
| TOPS | OC14 | 40567-80-4 | 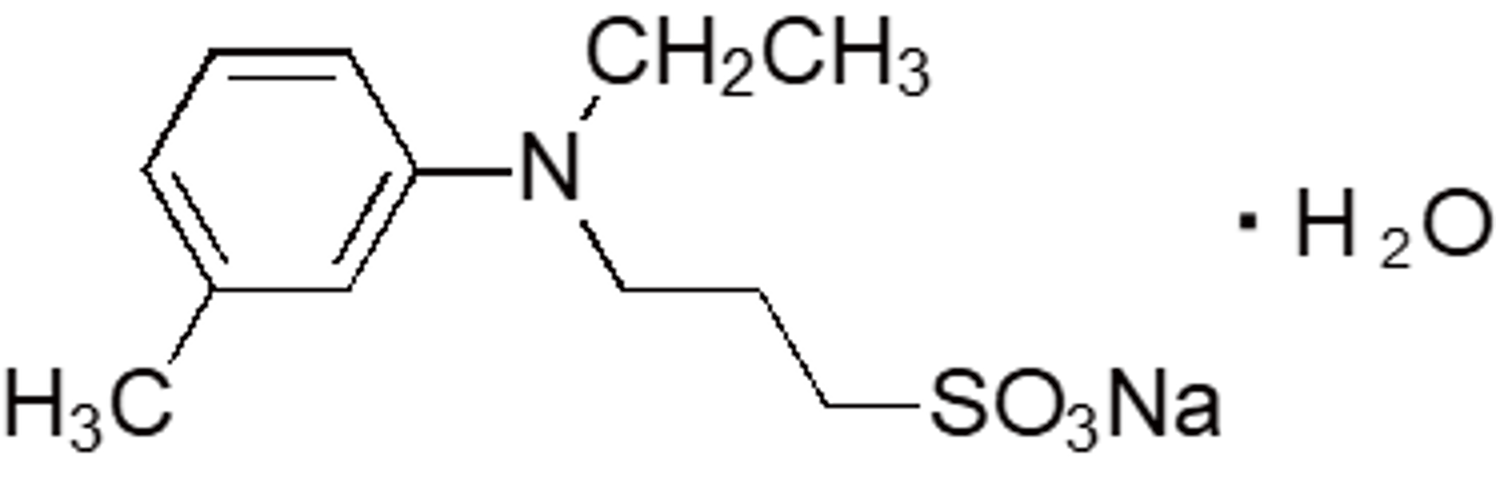 |
297.34 | 550 | 3.74 |
| ADPS | OC02 | 82611-88-9 | 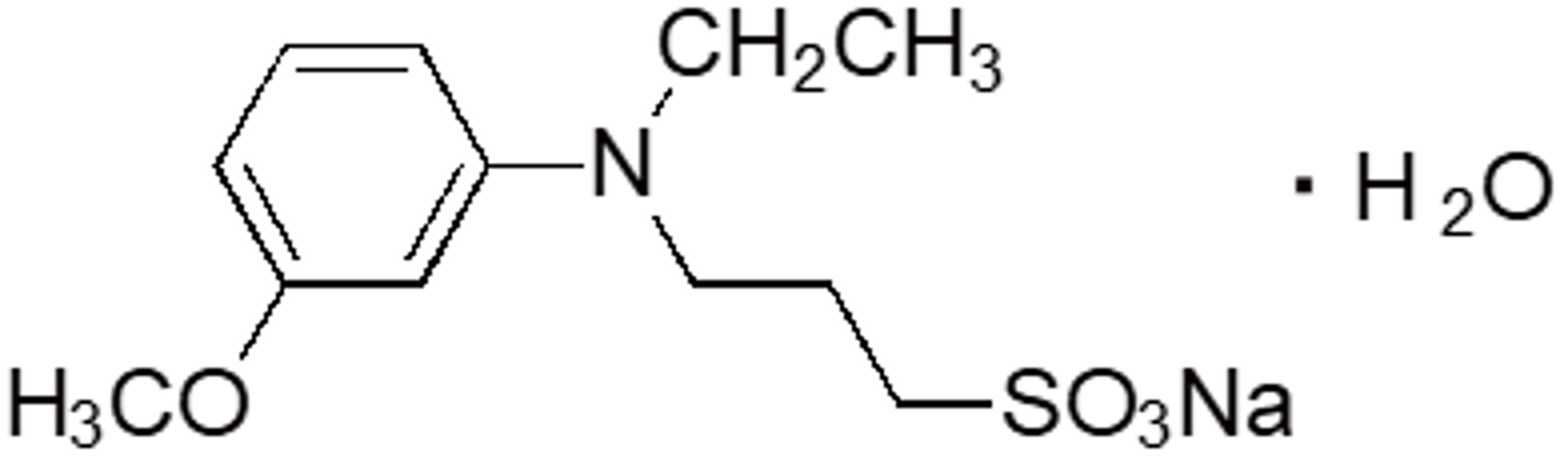 |
313.35 | 540 | 2.79 |
| ADOS | OC01 | 82692-96-4 | 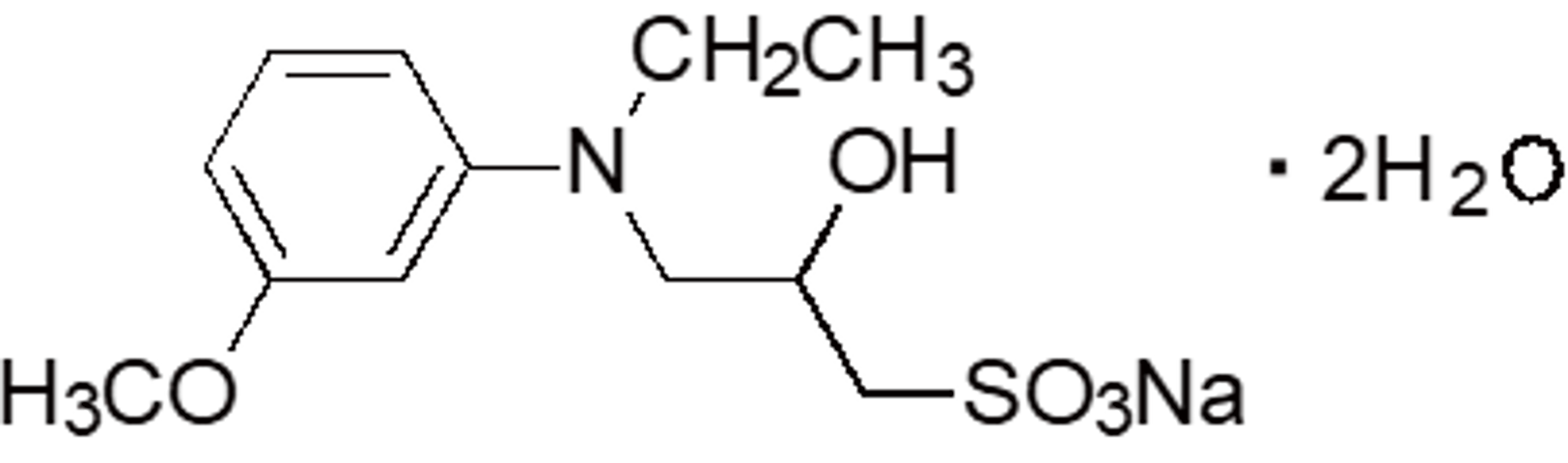 |
347.36 | 542 | 2.72 |
| MAOS | OC11 | 82692-97-5 | 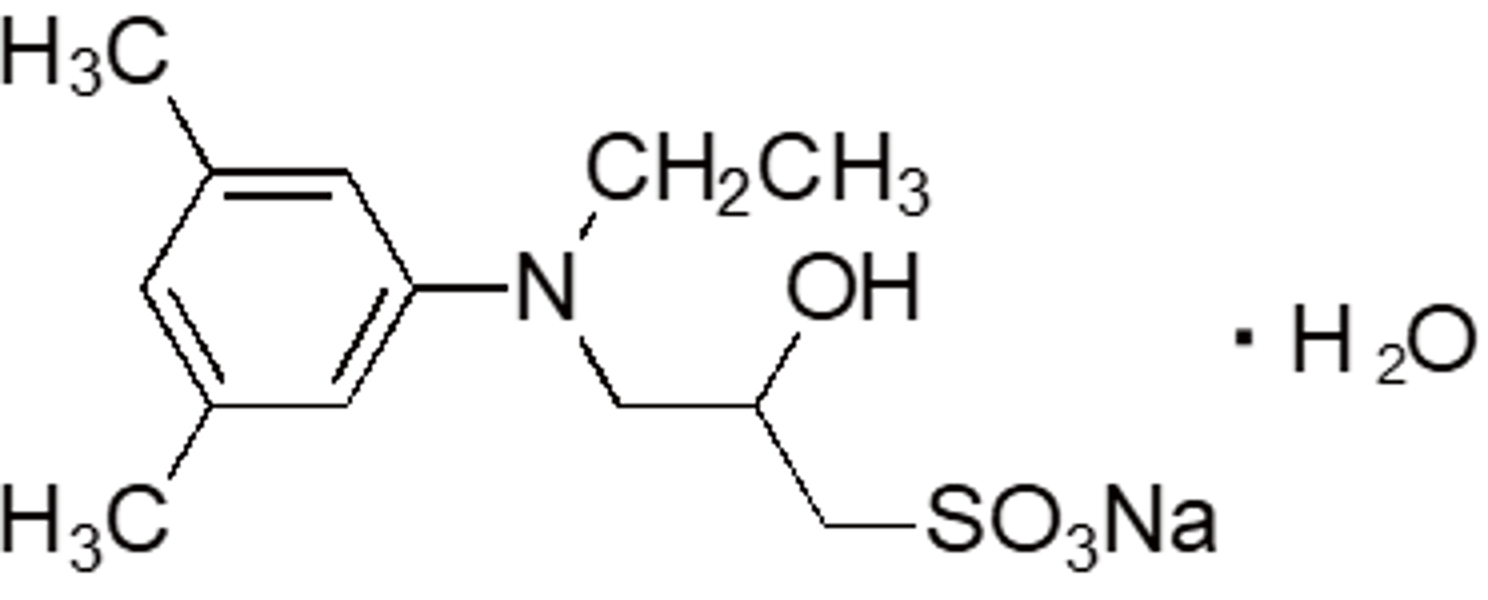 |
327.37 | 630 | 2.25 |
| DAOS | OC06 | 83777-30-4 | 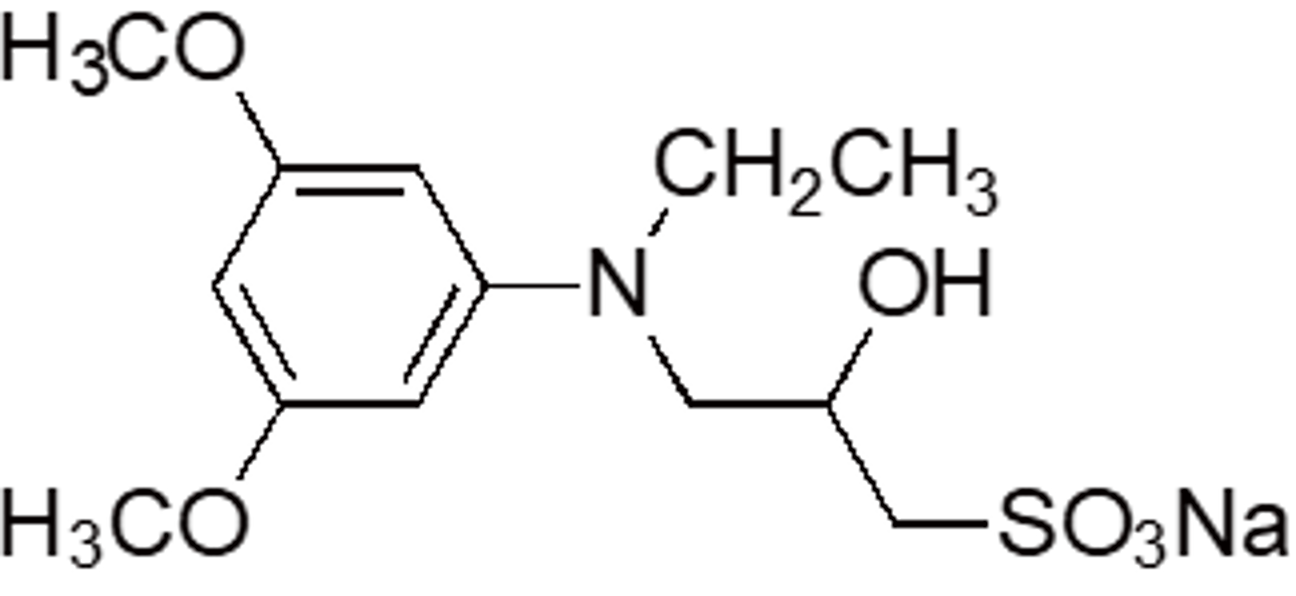 |
341.36 | 593 | 1.75 |
| HDAOS | OC08 | 82692-88-4 |  |
313.30 | 583 | 1.73 |
| MADB | OC21 | 209518-16-1 |  |
437.48 | 630 | 1.65 |
| EMSE | OC46 | - | 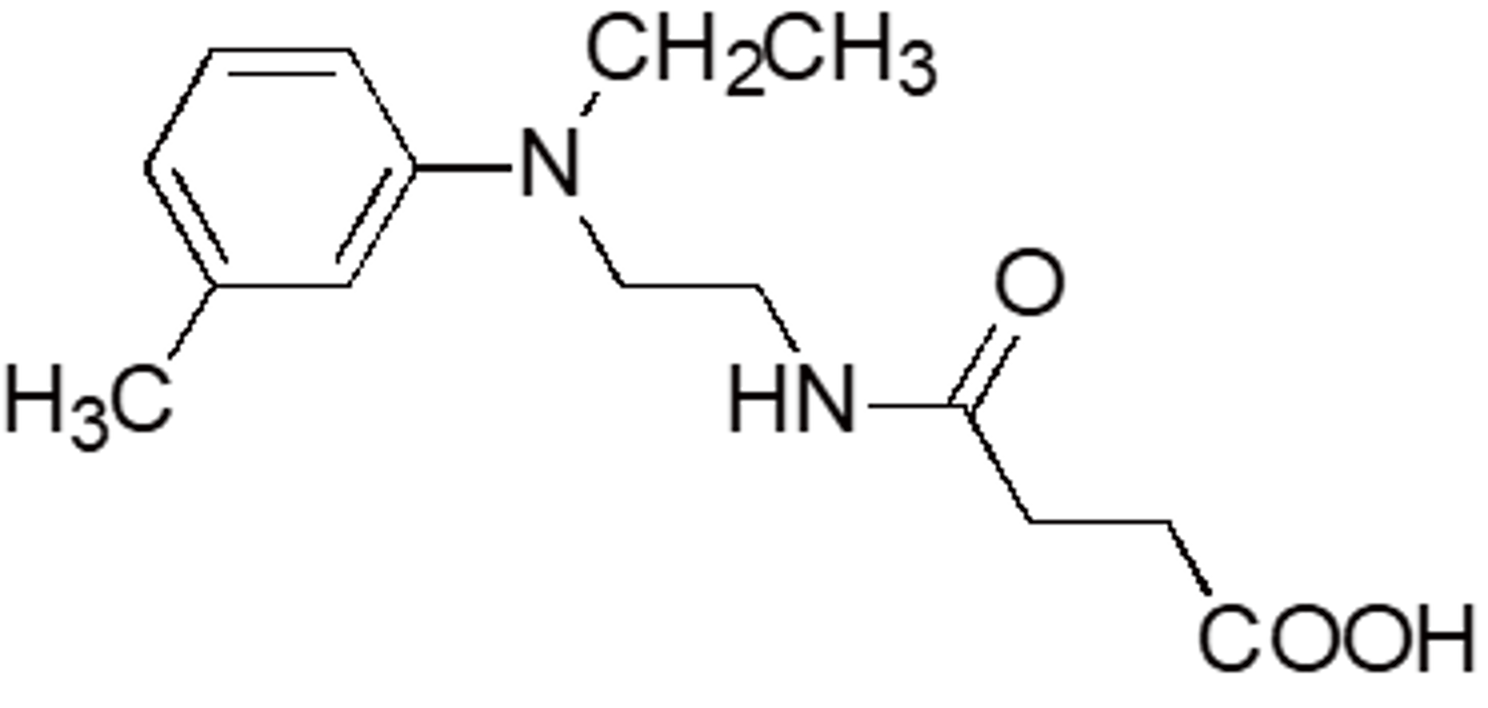 |
278.35 | 555 | - |
Redox Dyes
| Name | Code | CAS# | Chemical Structure | MW | Detecting Target |
|---|---|---|---|---|---|
| 5-Br-PAPS | B026 | 81608-06-2 (free acid) |
 |
537.34 | Zinc |
| 3,5-DiBr-PAESA | D041 | 100743-65-5 |  |
546.21 | Copper |
| Chlorophosphonazo-III | C010 | 1914-99-4 |  |
757.37 | Calcium |
| Nitroso-PSAP | N010 | 80459-15-0 | 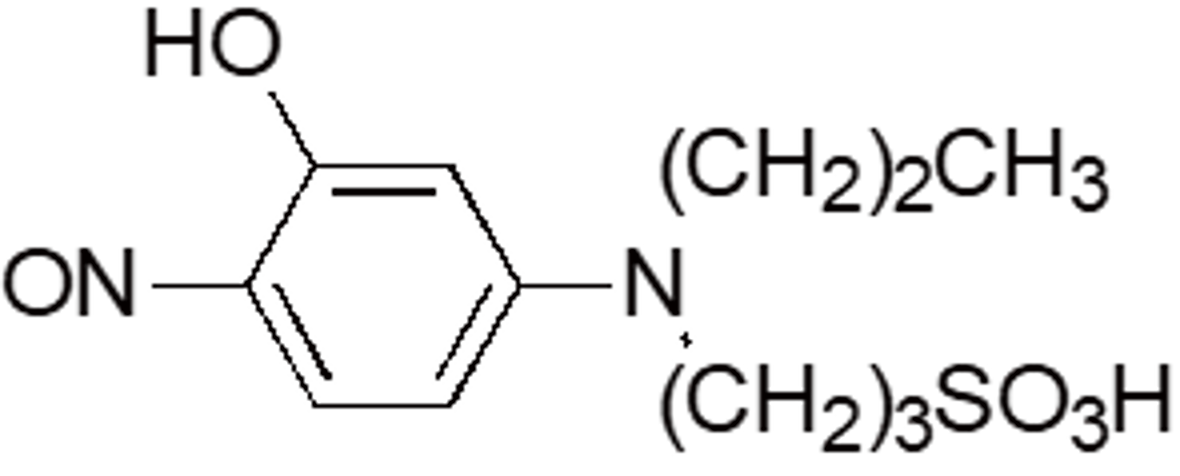 |
302.35 | Iron |
| Nitro-PAPS | N031 | 143205-66-7 (as anhydride) |
 |
503.45 | Zinc |
| PC | P004 | 2411-89-4 | 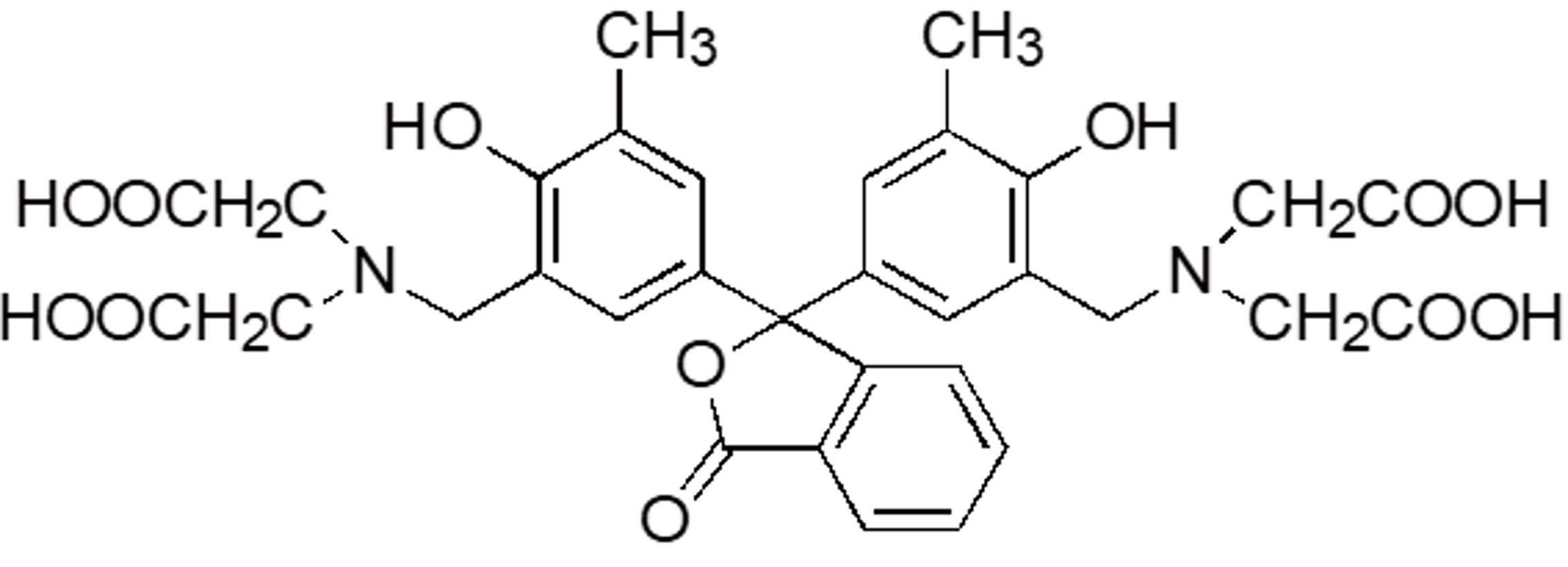 |
636.6 | Calcium |
| XB-I | X001 | 14936-97-1 |  |
513.5 | Magnesium |
| Glupa-C | G213 | 63699-78-5 | 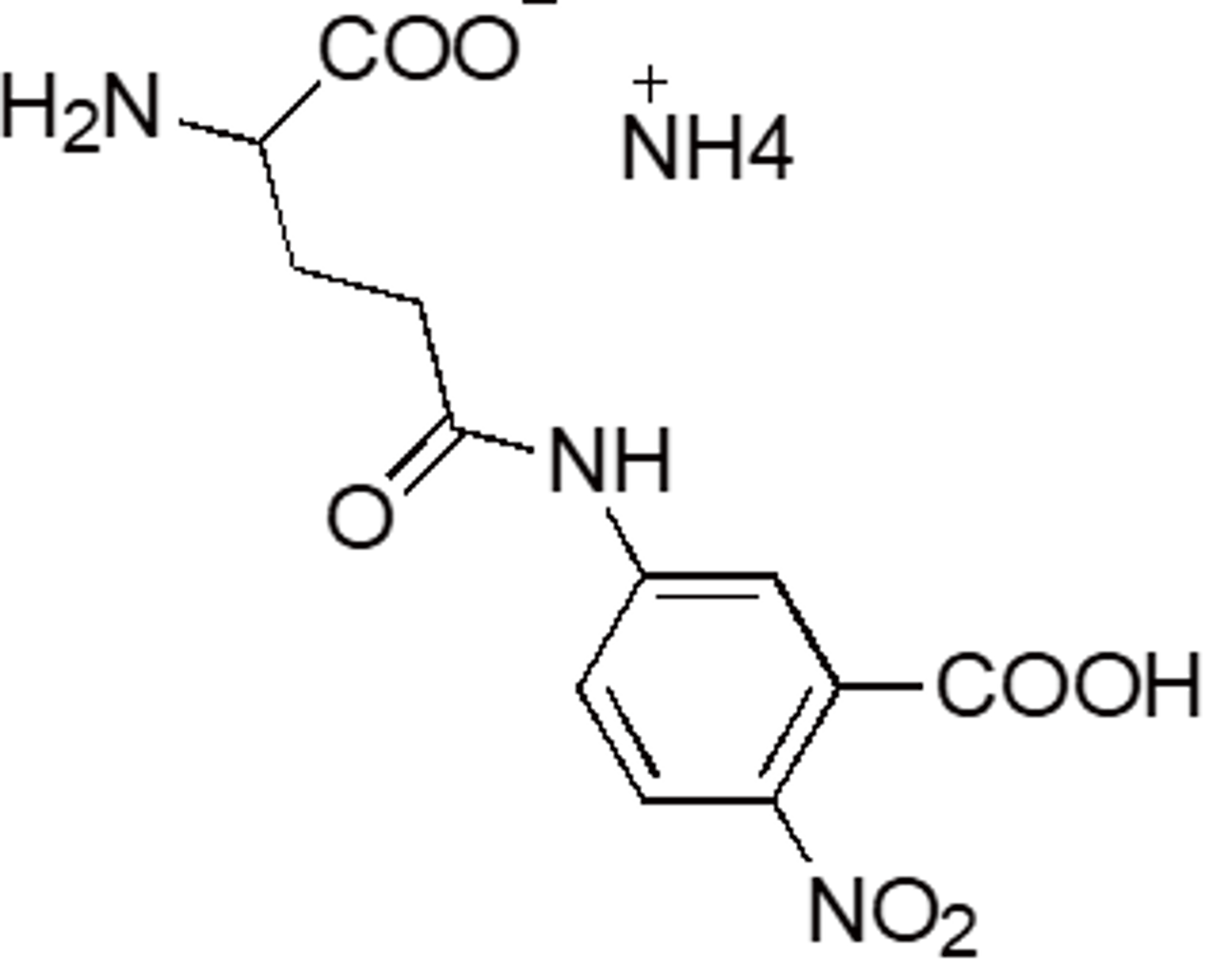 |
328.28 | Substrate for γ-glutamyltransferase |
| PNPP | N055 | 333338-18-4 (hexahydrate) |
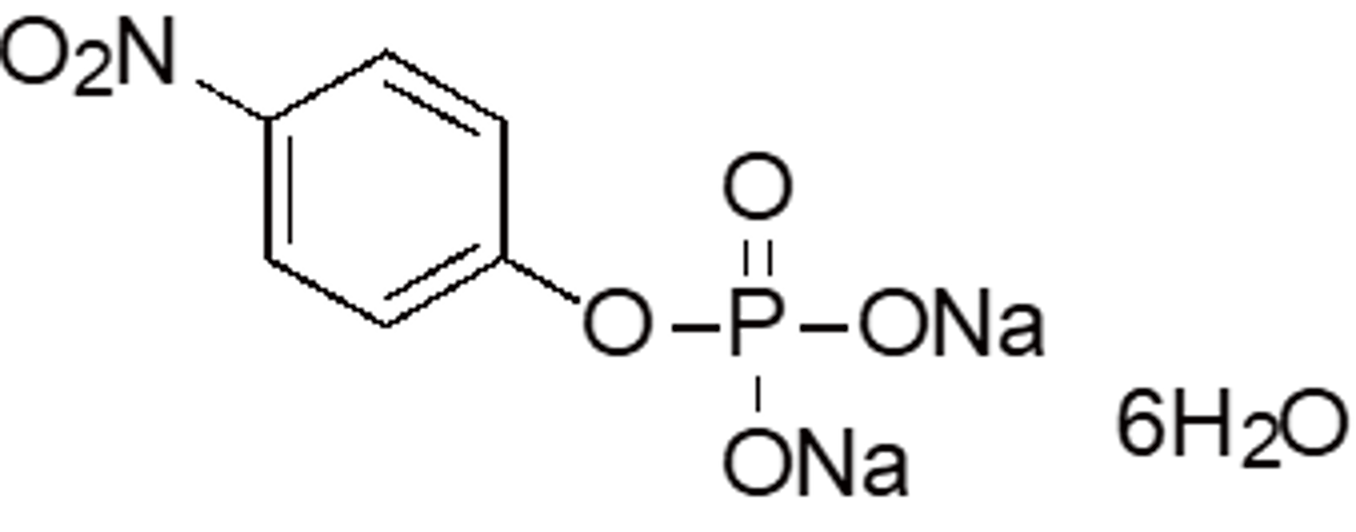 |
371.14 | substrate for alkaline phosphatase |
| Nitro-TB | N011 | 298-83-9 |  |
817.64 | Substrate for Dehydrogenase |

Buffers & Chelates
Good's Buffer
| Name | Code | Chemical Structure | MW | Solubility in Water (0°C, mol/l) |
pKa(20°C) | ΔpKa/°C | pH range |
|---|---|---|---|---|---|---|---|
| MES | GB12 | 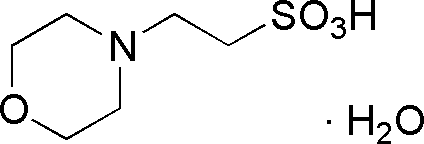 |
213.25 | 0.65 | 6.15 | -0.011 | 5.5 – 7.0 |
| Bis-Tris | GB05 | 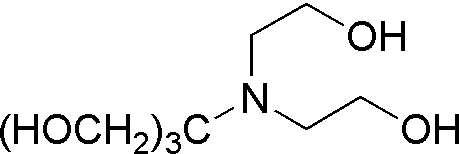 |
209.24 | >1.0 | 6.46 | – | 5.7 – 7.3 |
| ADA | GB02 | 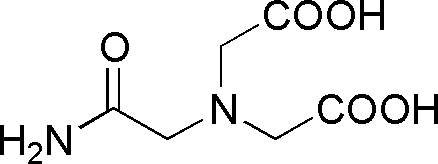 |
190.15 | insoluble | 6.6 | -0.011 | 5.8 – 7.4 |
| PIPES | GB15 | 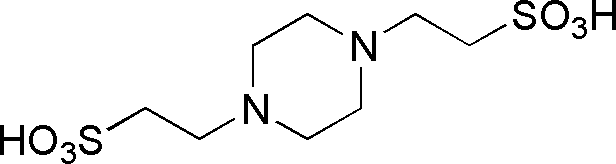 |
302.37 | insoluble | 6.8 | -0.0085 | 6.1 – 7.5 |
| PIPES sesquisodium |
GB25 | 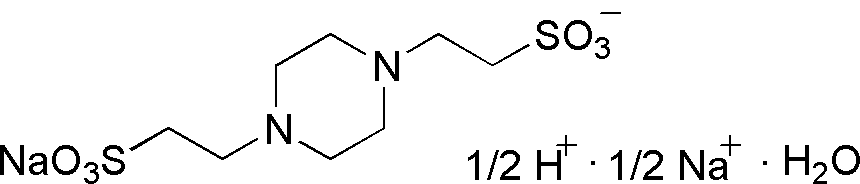 |
353.36 | >1.0 | 6.8 | -0.0085 | 6.1 – 7.5 |
| ACES | GB01 | 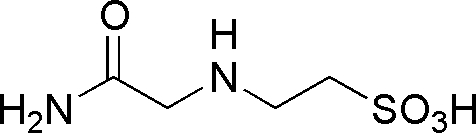 |
182.2 | 0.22 | 6.9 | -0.02 | 6.0 – 7.5 |
| MOPSO | GB14 | 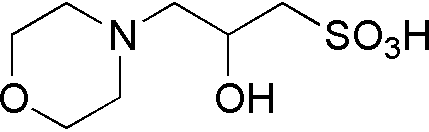 |
225.26 | 0.75 | 6.95 | – | 6.2 – 7.4 |
| BES | GB03 | 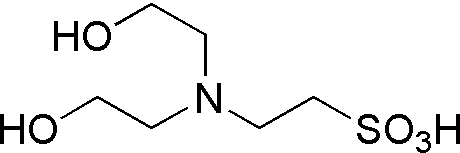 |
213.25 | 3.2 | 7.15 | -0.016 | 6.6 – 8.0 |
| MOPS | GB13 | 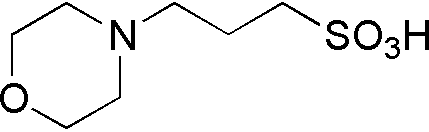 |
209.26 | 3 | 7.2 | -0.006 | 6.5 – 7.9 |
| TES | GB18 | 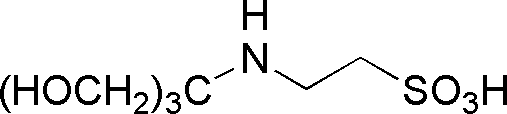 |
229.25 | 2.6 | 7.5 | -0.02 | 6.8 – 8.2 |
| HEPES | GB10 | 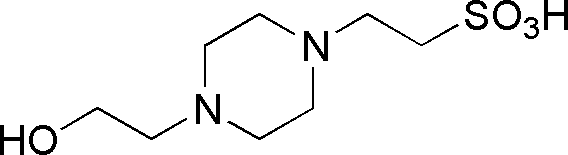 |
238.31 | 2.25 | 7.55 | -0.014 | 6.8 – 8.2 |
| TAPSO | GB20 | 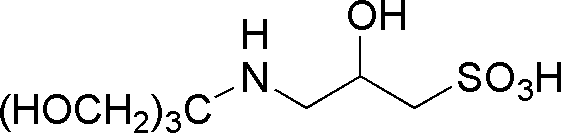 |
259.28 | 1 | 7.7 | – | 7.0 – 8.2 |
| POPSO | GB16 | 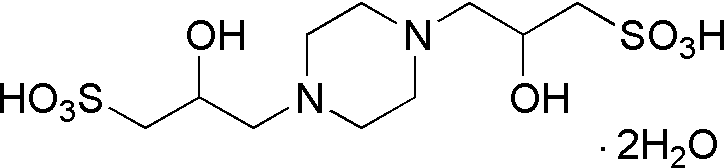 |
398.45 | insoluble | 7.85 | – | 7.2 – 8.5 |
| HEPPSO | GB11 | 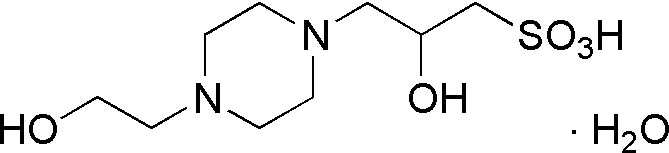 |
286.35 | 2.2 | 7.9 | – | 7.4 – 8.6 |
| EPPS | GB09 | 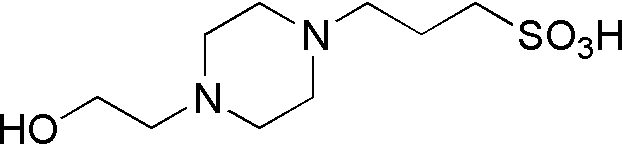 |
252.33 | 2.5 | 8 | -0.007 | 7.5 – 8.5 |
| Tricine | GB19 | 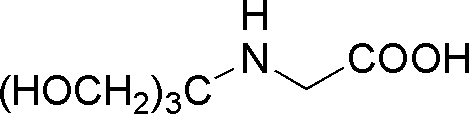 |
179.17 | 0.8 | 8.15 | -0.021 | 7.8 – 8.8 |
| Bicine | GB04 | 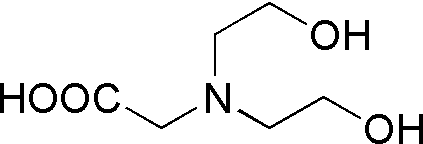 |
163.17 | 1.1 | 8.35 | -0.018 | 7.7 – 9.1 |
| TAPS | GB17 | 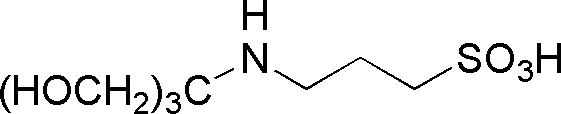 |
243.28 | >1.0 | 8.4 | – | 7.7 – 9.1 |
| CHES | GB07 | 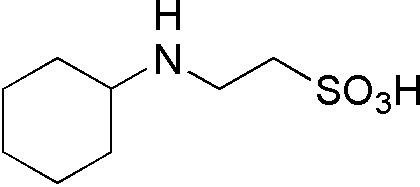 |
207.29 | 0.85 | 9.5 | -0.009 | 8.6 – 10.0 |
| CAPS | GB06 | 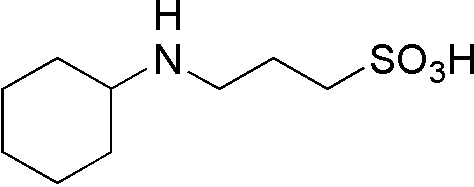 |
221.32 | 0.8 | 10.4 | -0.009 | 9.7 – 11.1 |
Metal Chelate
| Name | Code | Chemical Structure | MW | CAS# |
|---|---|---|---|---|
| EDTA-OH | E005 | 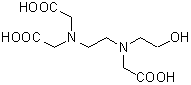 |
278.26 | 150-39-0 |
| GEDTA(EGTA) | G002 | 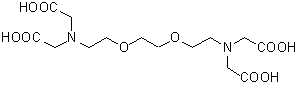 |
380.35 | 67-42-5 |
| 4H(EDTA・free acid) | H001 | 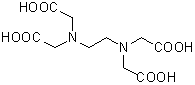 |
292.24 | 60-00-4 |
| 2K(EDTA・2K) | K001 | 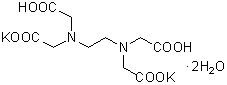 |
404.45 | 25102-12-9 |
| 3K(EDTA・3K) | K002 | 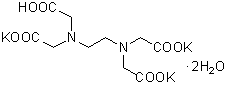 |
442.54 | 65501-24-8 |
| 2NA(EDTA・2Na) | N001 | 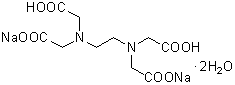 |
372.24 | 6381-92-6 |
| 3NA(EDTA・3Na) | N002 | 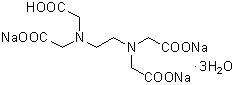 |
412.23 | 85715-60-2 |
| 4NA(EDTA・4Na) | N003 | 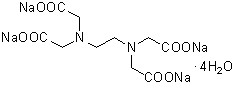 |
452.23 | 13235-36-4 |
| 2NH4(EDTA・2NH4) | N008 | 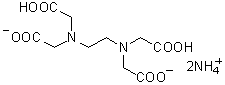 |
326.3 | 20824-56-0 |
| BAPTA | B019 | 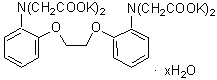 |
628.79 | 85233-19-8 (free acid) |
| CyDTA | C018 | 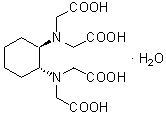 |
364.35 | 125572-95-4 |
| DTPA | D022 | 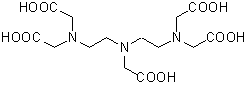 |
393.35 | 67-43-6 |
| HIDA | H006 | 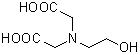 |
177.16 | 93-62-9 |
| NTPO | N030 | 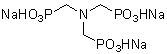 |
365 | 7611-50-9 |

Enzyme Substrates
| Name | Code | Chemical Structure | MW | CAS# |
|---|---|---|---|---|
| DAB | D006 | 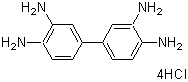 |
360.11 | 7411-49-6 |
| TMBZ | T022 | 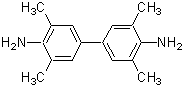 |
240.34 | 54827-17-7 |
| Glupa-C | G213 | 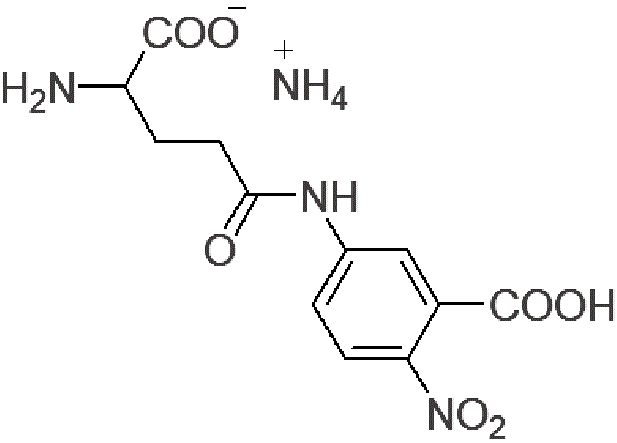 |
328.28 | 63699-78-5 |
| Disodium p-Nitrophenylphosphate | N055 | 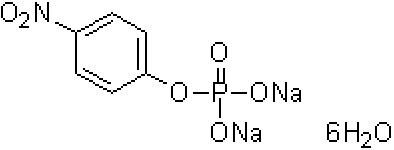 |
371.14 | 338-18-4 (hexahydrate) |

Cross-Linkers
Conjugation Kit
Amine or Thiol reactive Linkers
| Name | Code | Chemical Structure | MW | CAS# |
|---|---|---|---|---|
| EMCS | E018 | 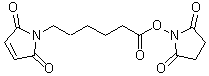 |
308.29 | 55750-63-5 |
| GMBS | G005 | 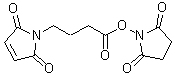 |
280.23 | 80307-12-6 |
| Sulfo-EMCS | S024 | 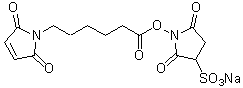 |
410.33 | 103848-61-9 (free acid) |
| Sulfo-GMBS | S025 | 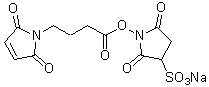 |
382.28 | 185332-92-7 |
| Sulfo-HMCS | S026 | 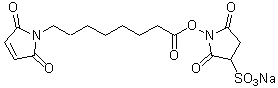 |
438.39 | 211236-35-0 |
| Sulfo-KMUS | S250 | 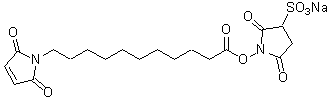 |
480.47 | 211236-68-9 |
| Sulfo-SMCC | S330 | 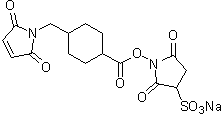 |
436.37 | 92921-24-9 |
| BS3 | B574 | 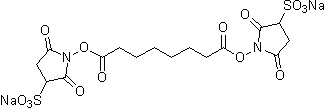 |
572.43 | 82436-77-9 (free acid) |
| Dithiobis(succinimidyl undecanoate) | D537 | 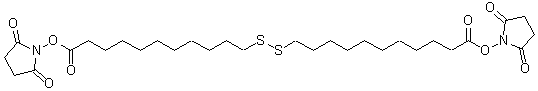 |
628.84 | 147072-47-7 |
| Dithiobis(succinimidyl hexanoate) | D539 | 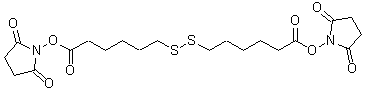 |
488.58 | 1083285-37-3 |
| DSP | D629 | 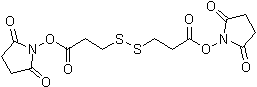 |
404.42 | 57757-57-0 |
| DTSSP | D630 | 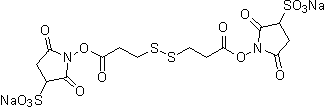 |
608.51 | 81069-02-5 (free acid) |
| SPDP | S291 | 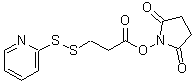 |
312.37 | 68181-17-9 |
| Sulfo-AC5-SPDP | S359 | 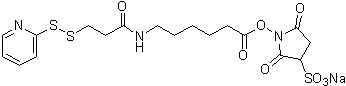 |
527.57 | 169751-10-4 |

Detergents
Nonionic
| Code | Name | Chemical Structure | MW | CAS# |
|---|---|---|---|---|
| BIGCHAP | B043 | 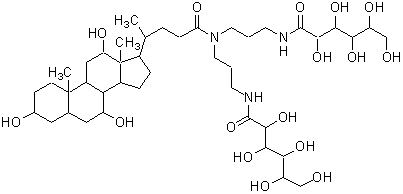 |
878.06 | 86303-22-2 |
| deoxy-BIGCHAP | D045 | 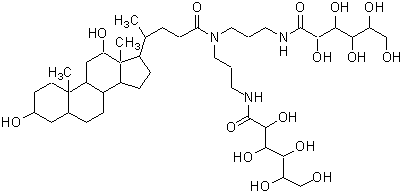 |
862.06 | 86303-23-3 |
| n-Dodecyl-β-D-maltoside | D316 | 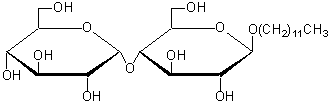 |
510.62 | 69227-93-6 |
| n-Decyl-β-D-maltoside | D382 | 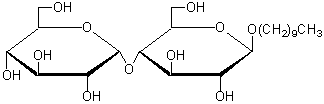 |
482.57 | 82494-09-5 |
| n-Heptyl-β-D-thioglucoside | H015 | 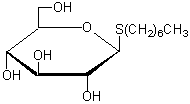 |
294.41 | 85618-20-8 |
| MEGA-8 | M014 | 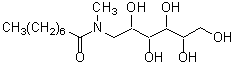 |
321.41 | 85316-98-9 |
| MEGA-9 | M015 | 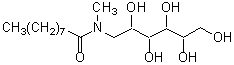 |
335.44 | 85261-19-4 |
| MEGA-10 | M016 | 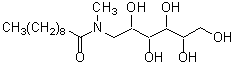 |
349.46 | 85261-20-7 |
| n-Nonyl-β-D-thiomaltoside | N373 | 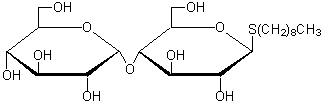 |
484.6 | 148565-55-3 |
| n-Octyl-β-D-glucoside | O001 | 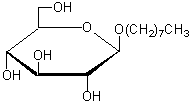 |
292.37 | 29836-26-8 |
| n-Octyl-β-D-thioglucoside | O003 | 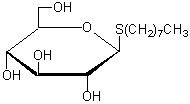 |
308.44 | 85618-21-9 |
| n-Octyl-β-D-maltoside | O393 | 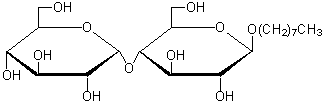 |
454.51 | 82494-08-4 |
| 3-Oxatridecyl-α-D-mannoside | O401 | 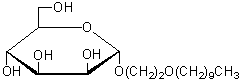 |
364.47 | 914802-92-9 |
| Trehalose C12 | T461 | 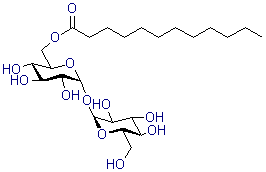 |
524.6 | 64622-91-9 |
| Trehalose C14 | T464 | 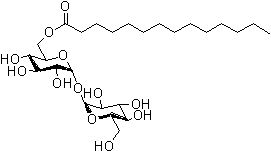 |
552.65 | 64622-92-0 |
Ionic
| Sodium cholate (purified) | C321 | 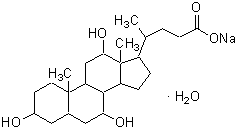 |
448.57 | 361-09-1 |
| Sodium deoxycholate (for protein crystallization) |
D520 | 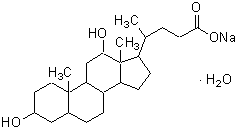 |
432.57 | 145224-92-6 |
Zwitterionic Type
| CHAPS | C008 | 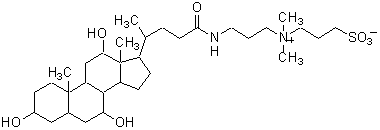 |
614.88 | 75621-03-3 |
| CHAPSO | C020 | 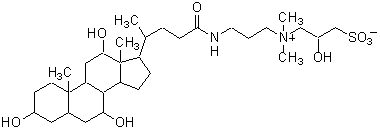 |
630.88 | 82473-24-3 |